Sex chromosome
| Part of a series on |
| Sex |
|---|
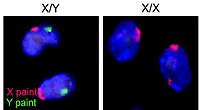 |
| Biological terms |
| Sexual reproduction |
| Sexuality |
| Sexual system |
Sex chromosomes (also referred to as allosomes, heterotypical chromosome, gonosomes, heterochromosomes,[1][2] or idiochromosomes[1]) are chromosomes that carry the genes that determine the sex of an individual. The human sex chromosomes are a typical pair of mammal allosomes. They differ from autosomes in form, size, and behavior. Whereas autosomes occur in homologous pairs whose members have the same form in a diploid cell, members of an allosome pair may differ from one another.
Nettie Stevens and Edmund Beecher Wilson both independently discovered sex chromosomes in 1905. However, Stevens is credited for discovering them earlier than Wilson.[3]
Differentiation
[edit]In humans, each cell nucleus contains 23 pairs of chromosomes, a total of 46 chromosomes. The first 22 pairs are called autosomes. Autosomes are homologous chromosomes i.e. chromosomes which contain the same genes (regions of DNA) in the same order along their chromosomal arms. The 23rd pair of chromosomes are called allosomes. These consist of two X chromosomes in females, and an X chromosome and a Y chromosome in males. Females therefore have 23 homologous chromosome pairs, while males have 22. The X and Y chromosomes have small regions of homology called pseudoautosomal regions.
An X chromosome is always present as the 23rd chromosome in the ovum, while either an X or Y chromosome may be present in an individual sperm.[4] Early in female embryonic development, in cells other than egg cells, one of the X chromosomes is randomly and permanently partially deactivated: In some cells, the X chromosome inherited from the mother deactivates; in other cells, it is the X chromosome inherited from the father. This ensures that both sexes always have exactly one functional copy of an X chromosome in each body cell. The deactivated X chromosome is silenced by repressive heterochromatin that compacts the DNA and prevents expression of most genes. This compaction is regulated by PRC2 (Polycomb Repressive Complex 2).[5]
Sex determination
[edit]
All diploid organisms with allosome-determined sex get half of their allosomes from each of their parents. In most mammals, females are XX, and can pass along either of their Xs; since males are XY they can pass along either an X or a Y. Females in such species receive an X chromosome from each parent while males receive an X chromosome from their mother and a Y chromosome from their father. It is thus the male's sperm that determines the sex of each offspring in such species.
However, a small percentage of humans have a divergent sexual development, known as intersex. This can result from allosomes that are neither XX nor XY. It can also occur when two fertilized embryo fuse, producing a chimera that might contain two different sets of DNA one XX and the other XY.[citation needed] It could also result from exposure, often in utero, to chemicals that disrupt the normal conversion of the allosomes into sex hormones and further into the development of either ambiguous outer genitalia or internal organs.[6]
There is a gene in the Y chromosome that has regulatory sequences that control genes that code for maleness, called the SRY gene.[7] This gene produces a testis-determining factor ("TDF"), which initiates testis development in humans and other mammals. The SRY sequence's prominence in sex determination was discovered when the genetics of sex-reversed XX men (i.e. humans who possess biological male-traits but actually have XX allosomes) were studied. After examination, it was discovered that the difference between a typical XX individual (traditional female) and a sex-reversed XX man was that the typical individuals lacked the SRY gene. It is theorized that in sex-reversed XX men, the SRY gets translocated to an X chromosome in the XX pair during meiosis.[8]
Other vertebrates
[edit]Diverse mechanisms are involved in the determination of sex in animals.[9] For mammals, sex determination is carried by the genetic contribution of the spermatozoon. Many lower chordates, such as fish, amphibians and reptiles, have systems that are influenced by the environment. Fish and amphibians, for example, have genetic sex determination but their sex can also be influenced by externally available steroids and incubation temperature of eggs.[10][11] In some reptiles, e.g. sea turtles, only the incubation temperature determines sex (temperature-dependent sex determination).
Plants
[edit]Many scientists argue that sex determination in plants is more complex than that in humans. This is because even flowering plants have a variety of mating systems, their sex determination primarily regulated by MADS-box genes. These genes code for proteins that form the sex organs in flowers.[12]
Plant sex chromosomes are most common in bryophytes, relatively common in vascular plants and unknown in ferns and lycophytes.[13] The diversity of plants is reflected in their sex-determination systems, which include XY and UV systems as well as many variants. Sex chromosomes have evolved independently across many plant groups. Recombination of chromosomes may lead to heterogamety before the development of sex chromosomes, or recombination may be reduced after sex chromosomes develop.[14] Only a few pseudoautosomal regions normally remain once sex chromosomes are fully differentiated. When chromosomes do not recombine, neutral sequence divergences begin to accumulate, which has been used to estimate the age of sex chromosomes in various plant lineages. Even the oldest estimated divergence, in the liverwort Marchantia polymorpha, is more recent than mammal or bird divergence. Due to this recency, most plant sex chromosomes also have relatively small sex-linked regions. Current evidence does not support the existence of plant sex chromosomes more ancient than those of M. polymorpha.[15]
The high prevalence of autopolyploidy in plants also impacts the structure of their sex chromosomes. Polyploidization can occur before and after the development of sex chromosomes. If it occurs after sex chromosomes are established, dosage should stay consistent between the sex chromosomes and autosomes, with minimal impact on sex differentiation. If it occurs before sex chromosomes become heteromorphic, as is likely in the octoploid red sorrel Rumex acetosella, sex is determined in a single XY system. In a more complicated system, the sandalwood species Viscum fischeri has X1X1X2X2 chromosomes in females, and X1X2Y chromosomes in males.[16]
Sequence composition and evolution
[edit]Amplification of transposable elements, tandom repeats especially accumulation of long tandom repeats (LTR) retrotransposones are responsible for plant sex chromosome evolution. The insertion of retrotransposons is probably the major cause of y-chromosome expansion and plant genome size evolution. Retrotransposones contribute in size determination of sex chromosomes and its proliferation varies even in closely related species. LTR and tandom repeats play dominant role in the evolution of S. latifolia sex chromosomes.[17] Athila is new family of retroelements, discovered in Arabidopsis thaliana, present in heterochromatin region only. Athila retroelements overrepresented in X but absent in Y while tandem repeats enriched in Y-chromosome. Some chloroplast sequences have also been identified in the Y-chromosome of S. latifolia. S. vulgaris has more retroelements in their sex chromosomes compare to S. latifolia. Microsatellite data shows that there is no significant difference between X and Y-chromosome microsatellites in both Silene species. This would conclude that microsatellites do not participate in Y-chromosome evolution. The portion of Y-chromosome that never recombine with X-chromosome faces selection reduction. This reduced selection leads to insertion of transposable elements and accumulation of deleterious mutation. The Y become larger and smaller than X due to insertion of retroelement and deletion of genetic material respectively. The genus Humulus is also used as model for the study of sex chromosomes evolution. Based on the phylogenetic topology distribution there are three regions on sex chromosomes. One region that stops recombining in the ancestor of H. lupulus, second that stops recombining in modern H. lupulus and the third region called pseudoautosomal region. H. lupulus is the rare case in plants in which Y is smaller than X, while its ancestor plant has the same size of both X and Y chromosomes. This size difference should be caused by deletion of genetic material in Y but that is not the case. This is because of complex dynamics like the larger size of X than Y-chromosome may be due to duplication or retrotransposition and size of Y remains same.[18]
Non-vascular plants
[edit]Ferns and lycophytes have bisexual gametophytes, so there is no evidence for sex chromosomes.[13] In the bryophytes, including liverworts, hornworts and mosses, sex chromosomes are common. The sex chromosomes in bryophytes affect what type of gamete is produced by the gametophyte, and there is wide diversity in gametophyte type. Unlike seed plants, where gametophytes are always unisexual, in bryophytes they may produce male, female, or both types of gamete.[19]
Bryophytes most commonly employ a UV sex-determination system, where U produces female gametophytes and V produces male gametophytes. The U and V chromosomes are heteromorphic with U larger than V and are frequently both larger than the autosomes. There is variation even within this system, including UU/V and U/VV chromosome arrangements. In some bryophytes, microchromosomes have been found to co-occur with sex chromosomes and likely impact sex determination.[19]
Gymnosperms
[edit]Dioecy is common among gymnosperms, found in an estimated 36% of species. However, heteromorphic sex chromosomes are relatively rare, with only five species known as of 2014. Five of these use an XY system, and one (Ginkgo biloba) uses a WZ system. Some gymnosperms, such as Johann's Pine (Pinus johannis), have homomorphic sex chromosomes that are almost indistinguishable through karyotyping.[16]
Angiosperms
[edit]Cosexual angiosperms with either monoecious or hermaphroditic flowers do not have sex chromosomes. Angiosperms with separate sexes (dioecious) may use sex chromosomes or environmental flowers for sex determination. Cytogenetic data from about 100 angiosperm species showed heteromorphic sex chromosomes in approximately half, mostly taking the form of XY sex-determination systems. Their Y is typically larger, unlike in humans; however there is diversity among angiosperms. In the Poplar genus (Populus) some species have male heterogamety while others have female heterogamety.[15] Sex chromosomes have arisen independently multiple times in angiosperms, from the monoecious ancestral condition. The move from a monoecious to dioecious system requires both male and female sterility mutations to be present in the population. Male sterility likely arises first as an adaptation to prevent selfing. Once male sterility has reached a certain prevalence, then female sterility may have a chance to arise and spread.[13]
In the domesticated papaya (Carica papaya), three sex chromosomes are present, denoted as X, Y and Yh. This corresponds with three sexes: females with XX chromosomes, males with XY, and hermaphrodites with XYh. The hermaphrodite sex is estimated to have arisen only 4000 years ago, post-domestication of the plant. The genetic architecture suggests that either the Y chromosome has an X-inactivating gene, or that the Yh chromosome has an X-activating gene.[20]
Medical applications
[edit]Allosomes not only carry the genes that determine male and female traits, but also those for some other characteristics as well. Genes that are carried by either sex chromosome are said to be sex linked. Sex linked diseases are passed down through families through one of the X or Y chromosomes. Since usually men inherit Y chromosomes, they are the only ones to inherit Y-linked traits. Men and women can get the X-linked ones since both inherit X chromosomes.[21]
An allele is either said to be dominant or recessive. Dominant inheritance occurs when an abnormal gene from one parent causes disease even though the matching gene from the other parent is normal. The abnormal allele dominates. Recessive inheritance is when both matching genes must be abnormal to cause disease. If only one gene in the pair is abnormal, the disease does not occur, or is mild. Someone who has one abnormal gene (but no symptoms) is called a carrier. A carrier can pass this abnormal gene to his or her children.[22] X chromosome carry about 1500 genes, more than any other chromosome in the human body. Most of them code for something other than female anatomical traits. Many of the non-sex determining X-linked genes are responsible for abnormal conditions. The Y chromosome carries about 78 genes. Most of the Y chromosome genes are involved with essential cell house-keeping activities and sperm production. Only one of the Y chromosome genes, the SRY gene, is responsible for male anatomical traits. When any of the 9 genes involved in sperm production are missing or defective the result is usually very low sperm counts and infertility.[23] Examples of mutations on the X chromosome include more common diseases such as the following:
- Color blindness or color vision deficiency is the inability or decreased ability to see color, or perceive color differences, under normal lighting conditions. Color blindness affects many individuals in the population. There is no actual blindness, but there is a deficiency of color vision. The most usual cause is a fault in the development of one or more sets of retinal cones that perceive color in light and transmit that information to the optic nerve. This type of color blindness is usually a sex-linked condition. The genes that produce photopigments are carried on the X chromosome; if some of these genes are missing or damaged, color blindness will be expressed in males with a higher probability than in females because males only have one X chromosome.
- Hemophilia refers to a group of bleeding disorders in which it takes a long time for the blood to clot. This is referred to as X-Linked recessive.[24] Hemophilia is much more common in males than females because males are hemizygous. They only have one copy of the gene in question and therefore express the trait when they inherit one mutant allele. In contrast, a female must inherit two mutant alleles, a less frequent event since the mutant allele is rare in the population. X-linked traits are maternally inherited from carrier mothers or from an affected father. Each son born to a carrier mother has a 50% probability of inheriting the X chromosome carrying the mutant allele.
- Fragile X syndrome is a genetic condition involving changes in part of the X chromosome. It is the most common form of inherited intellectual disability (mental retardation) in males. It is caused by a change in a gene called FMR1. A small part of the gene code is repeated on a fragile area of the X chromosome. The more repeats, the more likely there is to be a problem. Males and females can both be affected, but because males have only one X chromosome, a single fragile X is likely to affect them more. Most fragile-X males have large testes, big ears, narrow faces, and sensory processing disorders that result in learning disabilities.[25]
Other complications include:
- 46,XX testicular disorder of sex development, also called XX male syndrome, is a condition in which individuals with two X chromosomes in each cell, the pattern normally found in females, have a male appearance. People with this disorder have male external genitalia. In most people with 46,XX testicular disorder of sex development, the condition results from an exchange of genetic material between chromosomes (translocation). This exchange occurs as a random event during the formation of sperm cells in the affected person's father. The SRY gene (normally on the Y chromosome) is misplaced in this disorder, onto an X chromosome. Any person with an X chromosome that carries the SRY gene will develop male characteristics despite not having a Y chromosome.[26]
Evolution
[edit]Sex chromosomes evolve from standard pairs of autosomal chromosomes.[27] In a large number of organisms, the sex-determination systems presently observed are products of sex chromosome turnover. Sex chromosome turnover is a process defined as when the type of the sex chromosome changes as a product of a change in the identity of the sex-determining genes (such as by mutation) or by a change in their location.[28] In other cases, sex chromosomes may grow substantially with respect to their ancestral forms as a result of fusion events with autosomes, and autosome-sex chromosome fusions result in what are called neo-sex chromosomes. Five examples of this are now known in the songbird superfamily Sylvioidea.[29] There is one experimentally documented case of sex chromosome turnover occurring during a 30-year evolutionary experiment involving teleost fish (specifically the swordtails), in which hybridization experiments resulted in a translocation of the sex-determiner region of a sex chromosome into an autosome. This resulted in the autosome becoming a novel W sex chromosome.[30]
See also
[edit]- Fisher's principle
- Haldane's rule
- XY sex-determination system
- ZW sex-determination system
- X0 sex-determination system
References
[edit]- ^ a b "Allosome - Biology-Online Dictionary". www.biology-online.org. Archived from the original on 2018-02-11. Retrieved 2018-02-22.
- ^ "the definition of allosome". Dictionary.com. Archived from the original on 2016-02-04. Retrieved 2018-02-22.
- ^ Brush SG (June 1978). "Nettie M. Stevens and the discovery of sex determination by chromosomes". Isis. 69 (247): 163–172. doi:10.1086/352001. JSTOR 230427. PMID 389882. S2CID 1919033.
- ^ "How many chromosomes do people have?". Genetics Home Reference. U.S. National Library of Medicine. Archived from the original on 2013-04-02. Retrieved 2013-04-03.
- ^ Brockdorff N (November 2017). "Polycomb complexes in X chromosome inactivation". Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences. 372 (1733): 20170021. doi:10.1098/rstb.2017.0021. PMC 5627167. PMID 28947664.
- ^ Kucinskas L, Just W (2005). "Human male sex determination and sexual differentiation: pathways, molecular interactions and genetic disorders". Medicina. 41 (8): 633–640. PMID 16160410. Archived from the original on 2022-08-11. Retrieved 2022-03-29.
- ^ Graves, Jennifer A. Marshall (2015) Twenty-five years of the sex-determining gene Archived 2022-01-11 at the Wayback Machine, Nature 528, 343–344. https://doi.org/10.1038/528343a Retrieved 24 Jan 2022.
- ^ Hake L, O'Connor C (2008). "Genetic Mechanisms of Sex Determination". Scitable. Archived from the original on 2019-04-13. Retrieved 2019-10-24.
- ^ Hunter RH (March 1995). "Mechanisms of sex determination". Sex Determination, Differentiation and Intersexuality in Placental Mammals. Cambridge University Press. pp. 22–68. doi:10.1017/CBO9780511565274.003. ISBN 9780521462181. Archived from the original on 2018-06-10. Retrieved 2019-11-04.
- ^ Nakamura M (May 2009). "Sex determination in amphibians". Seminars in Cell & Developmental Biology. Environmental Regulation of Sex Dtermination in Vertebrates. 20 (3): 271–282. doi:10.1016/j.semcdb.2008.10.003. PMID 18996493.
- ^ Devlin RH, Nagahama Y (2002-06-21). "Sex determination and sex differentiation in fish: an overview of genetic, physiological, and environmental influences". Aquaculture. Sex determination and sex differentation in fish. 208 (3): 191–364. Bibcode:2002Aquac.208..191D. doi:10.1016/S0044-8486(02)00057-1. ISSN 0044-8486.
- ^ Ainsworth CC (1999). Sex determination in plants. Oxford, UK: BIOS Scientific Publishers. ISBN 0-585-40066-0. OCLC 50174640.
- ^ a b c Pannell JR (March 2017). "Plant Sex Determination". Current Biology. 27 (5): R191–R197. Bibcode:2017CBio...27.R191P. doi:10.1016/j.cub.2017.01.052. PMID 28267976.
- ^ Sun Y, Svedberg J, Hiltunen M, Corcoran P, Johannesson H (October 2017). "Large-scale suppression of recombination predates genomic rearrangements in Neurospora tetrasperma". Nature Communications. 8 (1): 1140. Bibcode:2017NatCo...8.1140S. doi:10.1038/s41467-017-01317-6. PMC 5658415. PMID 29074958.
- ^ a b Charlesworth D (April 2016). "Plant Sex Chromosomes". Annual Review of Plant Biology. 67 (1): 397–420. doi:10.1146/annurev-arplant-043015-111911. PMID 26653795.
- ^ a b Kumar S, Kumari R, Sharma V (April 2014). "Genetics of dioecy and causal sex chromosomes in plants". Journal of Genetics. 93 (1): 241–277. doi:10.1007/s12041-014-0326-7. PMID 24840848. S2CID 14956007.
- ^ Kralova T, Cegan R, Kubat Z, Vrana J, Vyskot B, Vogel I, et al. (2014). "Identification of a novel retrotransposon with sex chromosome-specific distribution in Silene latifolia". Cytogenetic and Genome Research. 143 (1–3): 87–95. doi:10.1159/000362142. PMID 24751661. S2CID 45809762.
- ^ Divashuk MG, Alexandrov OS, Kroupin PY, Karlov GI (2011). "Molecular cytogenetic mapping of Humulus lupulus sex chromosomes". Cytogenetic and Genome Research. 134 (3): 213–219. doi:10.1159/000328831. PMID 21709414. S2CID 23042305.
- ^ a b Renner SS, Heinrichs J, Sousa A (July 2017). "The sex chromosomes of bryophytes: Recent insights, open questions, and reinvestigations of Frullania dilatata and Plagiochila asplenioides: The sex chromosomes of bryophytes". Journal of Systematics and Evolution. 55 (4): 333–339. doi:10.1111/jse.12266.
- ^ VanBuren R, Zeng F, Chen C, Zhang J, Wai CM, Han J, et al. (April 2015). "Origin and domestication of papaya Yh chromosome". Genome Research. 25 (4): 524–533. doi:10.1101/gr.183905.114. PMC 4381524. PMID 25762551.
- ^ "Biological Basis of Heredity: Sex Linked Genes". Archived from the original on 2013-04-11. Retrieved 2013-04-07.
- ^ "Sex-linked recessive". PubMed. U.S. National Library of Medicine.
- ^ "Sex-Linked Traits". Heredity and genetics. Khan Academy. Archived from the original on 2013-04-08. Retrieved 2013-04-07.
- ^ "Hemophilia". PubMed Health. U.S. National Library of Medicine. Archived from the original on 2013-12-22. Retrieved 2017-11-01.
- ^ "Fragile X Syndrome - Symptoms, Diagnosis, Treatment of Fragile X Syndrome". NY Times Health Information. Archived from the original on 6 July 2013.
- ^ "46,XX testicular disorder of sex development". Genetics Home Reference. U.S. National Library of Medice. Archived from the original on 2013-03-30. Retrieved 2013-04-07.
- ^ Charlesworth D, Charlesworth B, Marais G (August 2005). "Steps in the evolution of heteromorphic sex chromosomes". Heredity. 95 (2): 118–128. doi:10.1038/sj.hdy.6800697. PMID 15931241. S2CID 10026903.
- ^ eLS (1 ed.). John Wiley & Sons, Ltd. 2001-05-30. doi:10.1002/9780470015902.a0028747. ISBN 978-0-470-01617-6. S2CID 208574438.
- ^ Sigeman H, Zhang H, Ali Abed S, Hansson B (December 2022). "A novel neo-sex chromosome in Sylvietta brachyura (Macrosphenidae) adds to the extraordinary avian sex chromosome diversity among Sylvioidea songbirds". Journal of Evolutionary Biology. 35 (12): 1797–1805. doi:10.1111/jeb.14096. PMC 10087220. PMID 36156325. S2CID 252543767.
- ^ Franchini P, Jones JC, Xiong P, Kneitz S, Gompert Z, Warren WC, et al. (December 2018). "Long-term experimental hybridisation results in the evolution of a new sex chromosome in swordtail fish". Nature Communications. 9 (1): 5136. Bibcode:2018NatCo...9.5136F. doi:10.1038/s41467-018-07648-2. PMC 6277394. PMID 30510159.
